
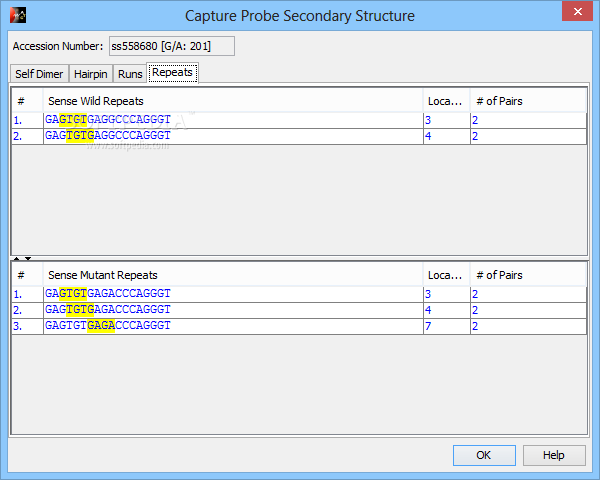
( B) Formation of non-target amplicon while joining of overlapping primer pairs into one multiplex reaction. L1.1 is a left primer of the first type for the first position L1.2 is a left primer of the second type for the first position R2.3 is a right primer of the third type for the second position etc. x i, x i+1, x i+2 are genome positions placed one by one. It is necessary because we can’t join two overlapping primer pairs into one multiplex reaction ( B). Such type of primer design gives more flexibility on the next steps, when primer pairs are combined into sets of primers that amplify the whole studied region (e.g. S3 Fig: ( A) Approach used by NGS-PrimerPlex, when for each position, the program calls primer3 to design three types of primers: so that the right primer was close to the studied position, the left primer was close to the studied position, and without any of these restrictions.
distribution among pools is restricted by a maximum of 15 primer pairs in one pool) ? –means that such functionality was claimed but the tool wasn’t acceptable ± –means that such functionality is partial (e.g. h–has graphical or web-interface yes/no–means that this tool includes or does not include such functionality, respectively. It can be useful for the detection of gene fusions. g–primer design for anchored PCR when the target region is amplified from one sequence-specific primer and one primer complemented to the adapter sequence. e–automatic primer distribution for several multiplex reactions, considering secondary structures and non-target amplicons that can be formed by primers from different pairs. d–checking primers for covering variable genome sites that contain high-frequent SNPs. c–checking primers for non-target hybridization and non-target amplification. b–primer design for all chosen genome regions. S1 Table: Comparison of NGS-PrimerPlex and other tools functionality: a–choosing genome regions to amplify by gene names and their parts.


 0 kommentar(er)
0 kommentar(er)
